Work Packages
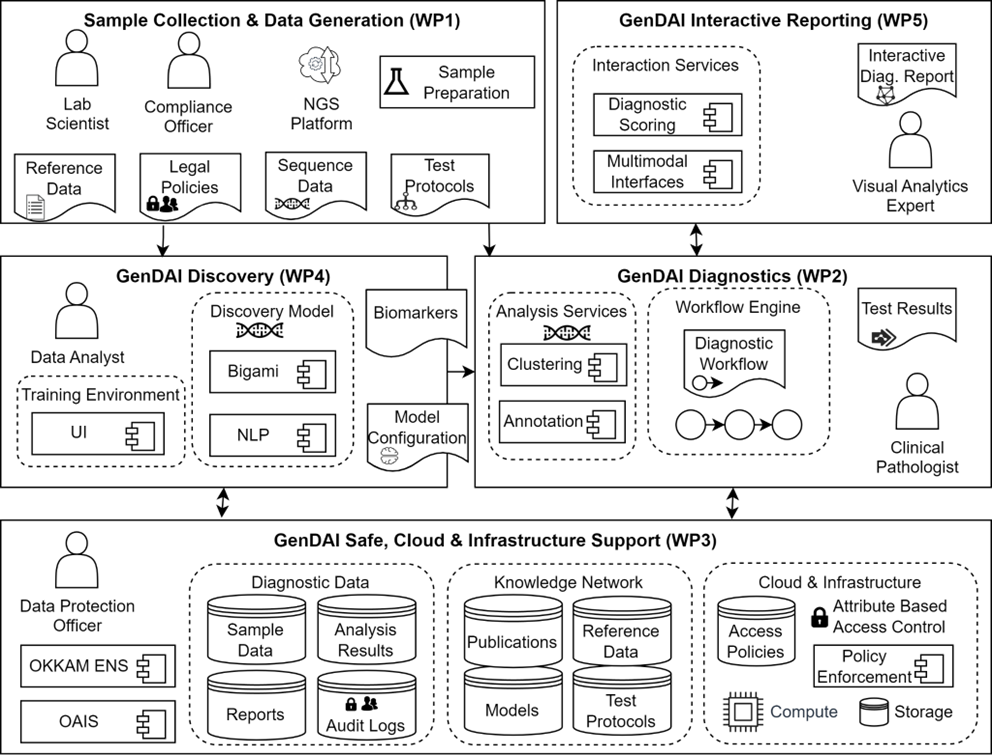
WP1: Clinical Requirements, Pre-analytics, sample Collection, Wet Lab Processing and Ethics (Lead: FTK)
Relevant functional and non-functional requirements are derived from a detailed analysis process and used for detailed modelling the system. These requirements and the models are used as input for the development of the components as described in the other work packages. Necessary steps for ethical and legal approval of sample collection are taken. Sample collection and provision using standard wet lab techniques and low-pass NGS are undertaken to create the datasets used for AI model training.
WP2: GenDAI Diagnostics – Advanced Genomics Data Processing Workflow (Lead: ICT)
The analysis and the reporting will be realised through an innovative Bioinformatics pipeline that supports the requirement of clinical diagnostics such as full reproducibility, auditing, automation, and standardisation to allow clinical validation in full compliance with applicable regulation. By integrating the components developed in other WPs, it will provide efficient analysis, annotation and classification to enable automated reporting about microbiome composition and healthy individual profiles.
WP3: GenDAI Safe & Cloud Computing Platform – Data Mgmt. & Long Term Archiving (Lead: SAP)
The GenDAI Safe & Cloud Computing Platform is designed for effective data management and long-term archiving, focusing on transparency and reproducibility. This effort encompasses developing a cloud-based infrastructure using Google Cloud services, integrating with the ENS for the assignment of PIDs to ensure traceability, and implementing dynamic, fine-grained access control policies supported by AI-based tools for policy authoring and auditing.
WP4: GenDAI Discovery – Biomarker prospection in the microbiome using Artificial Intelligence (Lead: MTU)
Focused in the development of novel AI methods for Metagenomics data Processing and Biomarkers identification, GenDAI will build and evaluate different AI models, including Random Forests, CNNs, DNNs and NLP, to pinpoint microbial components associated with IBS and potentially other diseases. GenDAI will also improve a previously established algorithm BiGAMi for feature selection in Metagenomic data, to explore its potential for biomarker identification and extraction in a custom model built by the user through split-learning.
WP5: GenDAI Interactive Reporting – Visual Data Analysis (Lead: UNIBA)
Through the visualisation of data produced by sequencing processes, GenDAI addresses several needs: 1) visualising the partial computation of the data in the cases where such computation goes beyond the perceived loss of interaction; 2) provide patients with interactive visualisations to improve the understandability of analysis result; 3) provide domain experts with advanced visual tools to perform comparison among different analysis and understand the available data.
WP6: Piloting and Validation (Lead: FTK)
Focused on an iterative process of deploying, evaluating, and refining prototypes for sample processing, classification, reporting and archiving. Feedback from clinical end users, including laboratory staff and pathologists at IMB, is integral to this cycle. Their insights guide the continuous improvement of each component, ensuring the final prototype is thoroughly validated and tailored to meet real-world clinical requirements.
WP7: Project Management (Lead: FTK)
Management of all aspects of the GenDAI project from an EC Grant Agreement enforcement standpoint and partners performance and satisfaction perspectives.
WP8: Dissemination, Communication & Exploitation (Lead: ICT)
Detailed programming and execution of the actions contained in the Dissemination, Communication and Exploitation Plan, aimed at maximising the scientific, social and economic impacts after project timeline and exploring and designing paths to exploitation and adoption of project outputs by healthcare services markets.
WP9: Ethics requirements (Lead: FTK)
This work package sets out the ‘ethics requirements’ that the project must comply with.